Researchers find that a new family of copper binding proteins is involved in fungal copper regulation
Researchers from INRAE, Marseille in France, Duke University School of Medicine, North Carolina in USA and University of Copenhagen (UCPH) discover that a new group of copper binding proteins is involved in fungal copper regulation. The research has just been published in two back to back articles in Nature Chemical Biology.
Many enzymes utilize metals for their biological function, other proteins bind metal ions to reduce their toxicity, or to bring these ions to where they are needed in the organism. Copper is a metal that is necessary for life, but is very reactive and therefore can generate chemical species that are toxic for living organisms.
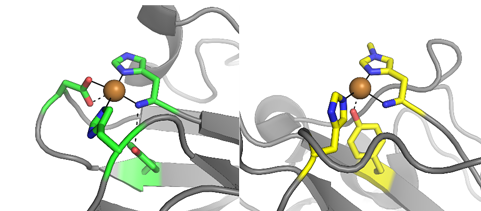
The copper binding sites of the protein X325 to the left and the cellulose cleaving LPMO TaAA9 to the right. Prepared by Kristian Frandsen.
The researchers have studied two related copper binding proteins from fungi found in very diverse habitats, one of them the human pathogen Cryptococcus. Initially they expected these proteins to be enzymes involved in degradation of polysaccharides, because their copper binding sites are very similar to the ones found in enzymes such as LPMOs (lytic polysaccharide monooxygenases).
Surprisingly, through studying their structure and properties, the researchers established that this is not their role. Instead, these important proteins are more likely to be involved in the regulation of copper ion content in the respective organisms. In Cryptococcus, the protein is involved in a pathway for copper uptake that drives brain colonization and thereby causes a type of meningitis.
Professor Leila Lo Leggio, from the Department of Chemistry (CHEM) at UCPH heading the group behind the structural studies, says:
It has been an exciting discovery journey. My group studies the atomic structure of proteins by X-ray crystallography, utilizing large X-ray producing facilities called synchrotrons. While investigating a family of proteins discovered by our coworkers in Marseille and Duke University, we discovered that their copper binding site was similar to the one found in our favorite enzymes LPMOs. LPMOs use copper to break bonds in polysaccharides from plant biomass that are difficult to degrade, thus they are important for exploiting waste biomass, to make new building blocks for synthesis or for biofuel production. However, the binding site in the new family of protein was a little different, with an extra chemical group binding to the copper ion, and other structural features just did not fit with this enzymatic role, which indeed has now been dismissed by our excellent co-workers at IGN, who have looked at the biochemical features.
Professor Katja S Johansen, head of the HOPE project at The Department of Geosciences and Natural Resource Management (IGN) at UCPH explains;
The study of the biochemical properties of these copper binding proteins was initiated before the crystal structure was solved. We repeatedly failed to detect catalytic activity in head-to-head comparative activity assays with well-described LPMOs. Certainly, this would be the case if there was something wrong with the samples, but that explanation was less likely when samples from two different laboratories and from different fungi give the same result. We are grateful for the opportunity to contribute to these studies and participate in this terrific collaboration. Fortunately, we are continuing the work and hope to be able to present further data in the future.
The research is published in two back to back articles in Nature Chemical Biology
https://www.nature.com/articles/s41589-019-0438-8
https://www.nature.com/articles/s41589-019-0437-9
The work carried out at IGN and CHEM was funded by Novo Nordisk Foundation. (NNF17SA0027704).
The groups involved in the research
Jean-Guy Berrin
INRAE Biodiversity and Biotechnology of Fungi
Faculté des Sciences de Luminy in Marseille
France
Dennis Thiele
Duke University School of Medicine
North Carolina
USA
Leila Lo Leggio
Department of Chemistry
University of Copenhagen
Katja S. Johansen
Department of Geosciences and Natural Resource Management
University of Copenhagen
Contact
Leila Lo Leggio
Professor
leila@chem.ku.dk
+45 35 32 02 95
Katja S. Johansen
Professor
ksj@ign.ku.dk
+45 31400588
